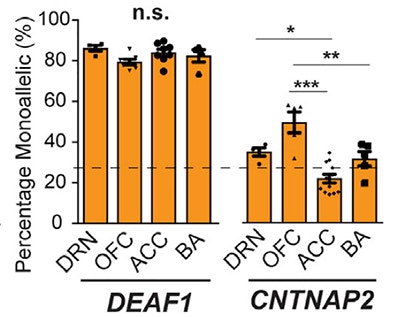
Epigenetic mechanisms can alter allelic gene expression patterns in the brain, and influence phenotypes and risk for developmental disorders. While genomic imprinting and random X-inactivation are two of the most well-known epigenetic mechanisms, less is known about whether other potential epigenetic mechanisms affect gene expression in the brain. In a new study, SFARI Investigator Christopher Gregg and colleagues developed a genomics strategy and statistical framework for performing genome-wide screens for diverse forms of non-genetic allelic expression effects in both mouse and primate brains. Using this methodology, the researchers uncovered developmental stage and cell-type specific non-genetic allelic effects that were not due to imprinting. Such effects were found to be particularly prevalent in the neonatal mouse brain. Assessments in macaque and postmortem human brains further indicated that such mechanisms can impact genes linked to neurodevelopmental disorders, including the expression of two autism risk genes, DEAF1 and CNTNAP2. These findings suggest that a broad array of epigenetic mechanisms should be considered, in addition to genetic risk variants, as potentially contributing to autism. Moreover, they provide a potential explanation for at least some of the phenotypic heterogeneity associated with autism-associated mutations, in that the impact of a mutation will depend on which allele is affected.

By clicking to watch this video, you agree to our privacy policy.
Reference(s)
Diverse non-genetic, allele-specific expression effects shape genetic architecture at the cellular level in the mammalian brain.
Huang W.C., Ferris E., Cheng T., Hörndli C.S., Gleason K., Tamminga C., Wagner J.D., Boucher K.M., Christian J.L., Gregg C.


