Previous studies of the genetic underpinnings of autism spectrum disorder (ASD) have revealed that many of the identified risk genes also confer substantial risk of intellectual disability and developmental delay. The overlap suggests that a combined genetic analysis of the three diagnoses is justifiable and can be used to boost statistical power.
To that end, SFARI Investigator Evan Eichler and colleagues carried out an integrated meta-analysis of de novo mutations from individuals with at least one of these related neurodevelopmental disorders (NDD). The exome sequencing data they examined were from 10,927 individuals, including the Simons Simplex Collection. Combining the results of two different statistical models, they identified 253 genes with an excess of missense and/or likely gene-disruptive (LGD) mutations, of which 124 reached exome-wide significance. This represents the most expansive gene list yet published for neurodevelopmental disorders, which also highlights a number of candidates for drivers of the effects of NDD-associated copy number variants.
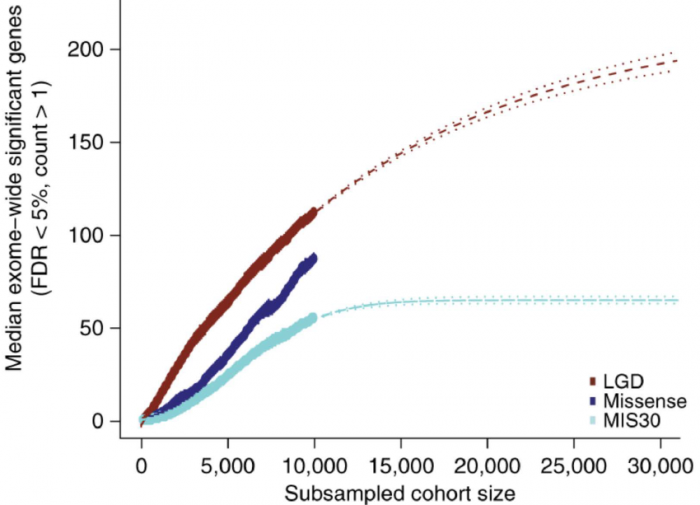
Among the other findings are a demonstration of biological convergence of the implicated genes on a small number of functional modules, including regulation of transcription, synaptic signaling and kinase-dependent signal transduction, as well as expression bias of these genes in mid-fetal cortex, striatum and amygdala. Of particular note, the authors estimate that a tripling of sample size will identify just over 200 exome-wide significant genes implicated by LGD mutation, beyond which there may be diminishing returns.
Reference(s)
Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity.
Coe B.P., Stessman H., Sulovari A., Geisheker M.R., Bakken T.E., Lake A.M., Dougherty J., Lein E.S., Hormozdiari F., Bernier R., Eichler E.


