While hundreds of genes conferring risk for autism spectrum disorders (ASD) have now been identified, the mechanisms whereby pathogenic variants disrupt behavior remain largely unknown. To shed light on the biology of genes conferring ASD risk, numerous animal models carrying ASD-relevant mutations have been created. Though the majority of models make use of rodent systems, fish, fruit flies and worms have also been employed. Of the non-mammalian systems, zebrafish (Danio rerio) has attracted increasing attention, and, in the past few years, SFARI has funded several projects using this model system to study molecular and circuit mechanisms relevant to autism.
To learn more about current and potential applications of zebrafish as an experimental system for the study of autism and related neurodevelopmental disorders, SFARI organized a workshop on January 26, 2018. This workshop brought together researchers from different academic backgrounds and with experience in both rodent and fish models as well as human genetics. The aim of the meeting was to discuss the current status of zebrafish-based research, with a focus on questions regarding the construct validity of mutant lines, the visualization of circuit development and function, and translational prospects.
A summary of the main topics discussed is presented below.
Mutant zebrafish lines: Opportunities and challenges
Following brief introductions on autism genetics by SFARI investigator Joseph Buxbaum (Icahn School of Medicine at Mount Sinai) and SFARI senior scientist Alan Packer, the workshop kicked off with presentations discussing opportunities and challenges of using zebrafish mutant lines— a first step toward understanding how genes impact behavior.
The first speaker of the day, Peixin Zhu (Novartis/Harvard University), studied zebrafish collective behaviors with the goal of characterizing the genetic architecture of social behaviors. In his research, he used machine learning algorithms to create ‘ethograms’ of zebrafish collective behavior, based on their different movement states during shoaling. By comparing ethograms of wildtype and mutant individuals, he identified classes of genes associated with particular states (e.g., alignment, aggregation, dispersion, transitioning), which suggest different genetic contributions to different group behaviors.
Although over 400 million years of evolution separate humans and zebrafish, roughly 70 percent of zebrafish and human genes are orthologous. In his presentation, SFARI investigator Nicholas Katsanis (Duke University) argued that this genetic similarity with humans, combined with cheaper housing costs and faster reproduction rates than rodents, makes zebrafish an ideal experimental system to study (pathogenic) variation of the human genome. Illustrating this point, Katsanis described several cases from the Duke Task Force for Neonatal Genomics in which in vivo assays with zebrafish mutants were employed to study some of the pathological manifestations observed in the human participants in this project. Using zebrafish — or any other experimental animal system — to study the effects of disease-associated genes may help understand signaling pathways or mechanisms that are conserved in evolution.
The workshop also discussed some of the challenges involved in generating zebrafish mutant lines. While genetic manipulations are relatively easy to implement in zebrafish, and novel techniques (e.g., CRISPR) have increased the range of experimental possibilities, it is vital that mutant lines are verified experimentally to ensure that they were generated correctly. This issue was recently raised by studies claiming that, in some mutants, there was no actual loss of the RNA or protein encoded by the gene. Accordingly, Steven Farber (Carnegie Institution for Science) presented examples from his research on lipid metabolism, describing a number of possible scenarios by which genes could ‘escape’ mutations such that significant amounts of mRNA were still produced, including exon skipping, cryptic splice sites, nonsense-associated alternative splicing, or the use of alternative initiation codons. Farber’s research has found this so-called ‘genomic compensation’ to be a more common problem than previously appreciated2. Following his presentation, the group brainstormed methods to facilitate mutant validation and considered ways of generating mutants in which these issues would be less likely to occur.
High-throughput phenotyping in vivo
Moving from the generation and characterization of mutant lines to potential applications, a number of presentations then discussed translational opportunities, with a focus on drug testing and discovery.
By clicking to watch this video, you agree to our privacy policy.
Zebrafish can be studied at both the adult and larval stages. While adult fish have a larger behavioral repertoire than larvae, the small size of larvae offers the opportunity to test many thousands of individuals (and conditions) in standard experimental setups (e.g., 96 well plates). The large scale of these experiments (Video 1), combined with the fact that zebrafish behavioral analysis can now be completely automated [using commercially available systems such as the Vertebrate Automated Screening Technology (VAST) BioImager3], allows for high-throughput phenotyping—an advantage over rodent models, which typically employ only a small number of individuals and few behavioral tests.
High-throughput phenotyping not only provides a multidimensional assessment of behavior, but it also makes it possible to conduct phenotype-driven pharmacological screenings, as opposed to conducting target-driven methodologies4. Using this approach, Randall Peterson (University of Utah) examined the effects of early exposure to chemical compounds on the development of typical zebrafish social behavior later in life. As many more behaviors can be assessed in larvae (e.g., threat response, motility5), Peterson recommended taking an approach that combines data from multiple behavioral assays, compounds and mutants into databases, so these can be used as references in chemical screenings. He also argued that one effective way of using these screens to study ASD would be to compare response to chemical compounds between wildtype and mutant zebrafish carrying autism-relevant mutations; drugs that suppress or revert abnormal phenotypes to baseline could then be identified.
SFARI Investigator Ellen Hoffman (Yale University) has taken such an approach in her research (Figure 1). Specifically, she described results from a recent study6 in which she conducted pharmaco-behavioral profiling of zebrafish with loss-of-function in cntnap2, a high-confidence autism risk gene. After establishing that the mutants exhibited increased night hyperactivity relative to the wildtype, she screened for compounds that correlated and anti-correlated with this phenotype; she found that estrogen-containing drugs best repressed this behavior. Hoffman was awarded a SFARI Pilot Award in 2015 to apply this approach to the study of additional zebrafish lines with mutations in high-confidence ASD genes. This year, she was awarded a SFARI Research Award in which she will investigate whether the mutant phenotypes and drug anti-correlations identified in zebrafish translate to mouse models of ASD.

As drugs generally have multiple target and off-target effects, and therapeutic interventions may require pharmacological combinations, high-throughput chemical screens in zebrafish are likely to be key to testing all possible targets quickly and cost effectively. To this end, Mehmet Fatih Yanik (ETH Zurich) developed an experimental approach to assess the effects of chemical compounds directly on brain activity patterns. He used zebrafish carrying mutations in scn1lab, a gene that is orthologous to the family of sodium voltage-gated channel alpha subunit genes in humans (e.g., SCN1A, SCN2A) to investigate epileptic forms and behaviors relevant to ASD. By recording local field potentials in zebrafish scn1lab mutants, Yanik showed that brain activity patterns can help identify drugs with the greatest efficacy in reducing seizures and other abnormal behavioral metrics7.
A key discussion point related to these behavioral screens was the notion of face validity versus construct validity. The group agreed that relying on face validity (e.g., behaviors in zebrafish that might, on their face, look like behavioral aspects of ASD) was problematic, as such behaviors might be mediated by different underlying mechanisms in different species. Instead, SFARI’s priority is to study zebrafish lines with construct validity (e.g., those with mutations in genes known to confer high-risk for ASD in humans). Within these lines, behavioral phenotypes may still serve as a useful readout (e.g., for drug-screening assays), but the presence of altered social behavior does not confer relevance to autism per se. This issue was also recently highlighted in a report by the National Advisory Mental Health Council Workgroup on Genomics8, which emphasized the fundamental value of animal studies for basic science investigations, rather than as ‘models’ of human disease — a recommendation that went as far as to suggest replacing the terms ‘animal model’ or ‘disease model’ with ‘experimental systems.’
Visualizing circuit development, structure and function
In addition to providing opportunities for high-throughput screening assays, zebrafish provide a real-time window into the development of neural circuits and their structure and function.
SFARI investigator Ethan Scott (University of Queensland) described the application of whole-brain calcium imaging to understanding neural function at the single-cell level in the zebrafish larvae sensory system. Scott showed that zebrafish neurons can be clustered based on stimulus-specific responses (e.g., different auditory frequencies, waterflow types) and aims to study how these circuits may be affected in zebrafish carrying ASD-relevant mutations. In the past few years, Scott has received two awards from SFARI, an Explorer Award to examine the development of Purkinje cells in several mutants (i.e., tsc1a, nrx1a, auts2, scn1lab, grik2 and gabrb3) and, more recently, a Pilot Award to use time-lapse imaging of developing neurons in zebrafish carrying mutations for additional ASD risk genes.
Also using whole-brain imaging, Harry Burgess (Eunice Kennedy Shriver National Institute of Child Health and Human Development) focused on zebrafish brain structure and illustrated the use of voxel-based morphometry and deformational analyses9, 10 to assess neurodevelopmental abnormalities in cell type (e.g., GABA-expressing neurons) number and density, and neuroanatomical region size. Both Scott’s and Burgess’ presentations underscored another advantage of using zebrafish, namely, the capability to visualize function and structure at high resolution in vivo, in the intact brain and in an unbiased fashion.
Zebrafish may also allow for the study of developmental mechanisms at multiple scales, from molecular through cellular to circuit level. Joseph Fetcho (Cornell University) argued that this approach would be particularly useful to study ASD, as processes that are purportedly disrupted in this disorder (e.g., patterning and synaptogenesis, neurotransmission, homeostatic balance and connectivity) can be effectively observed in zebrafish. In fact, because zebrafish larvae are optically transparent, many of those processes can be directly visualized, which facilitates tracking patterns and timing of typical and atypical development.
“We can see precisely when and how neural development goes awry,” said Burgess.
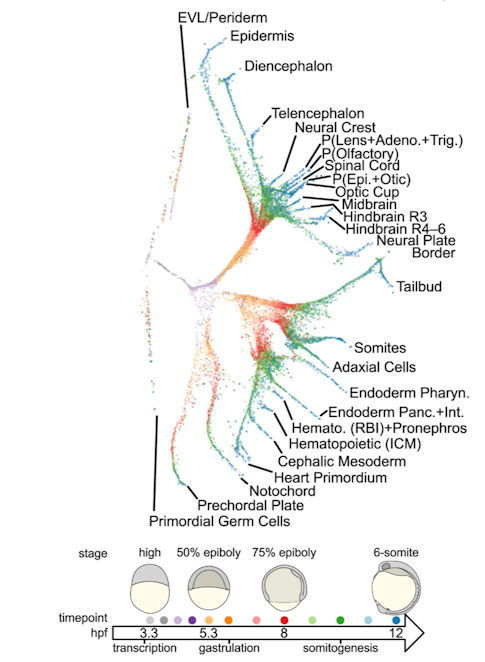
Finally, using zebrafish embryos, Alex Schier (Harvard University) showed that single cell RNA sequencing can be used to determine each cell’s identity11. This methodology is useful not only to cluster cells by type (e.g., neuron, progenitors, blood, vasculature, glia) and map them onto the brain12, but also to build ‘trees’ of cell transcriptional trajectories (Figure 2). Moreover, via genome editing, cells can be marked to reconstruct the lineage relationships of their descendants13. Outcomes of this research include a fine-grained, cellular and temporal characterization of zebrafish embryogenesis, which may help build gene-expression atlases, as well as a better understanding of the factors that determine cell fate. The same approach can also be used to define disease progression and disease states.
Final remarks
Offering an attractive compromise between system tractability, behavioral complexity, genetic similarity with humans and housing costs, zebrafish provide a promising experimental tool. The manifold uses for drug discovery and understanding the genetic and neural bases of human behavior and disease make them particularly interesting to study neurodevelopmental disorders, including ASD.
“Not using zebrafish would be dumb,” said Fetcho.
There are, however, issues the field is still grappling with, such as generating experimentally valid mutant lines and the notion of face validity. Furthermore, it is worth pointing out that although many mechanisms are evolutionarily conserved and human and zebrafish genes are in part orthologous, millions of years of evolution have contributed to shaping human and zebrafish species-specific neural adaptations. Thus, the type of research questions that can be addressed by this species — and, indeed, any other animal experimental system — and be relevant for ASD needs to be carefully considered.
How SFARI might leverage the potentials of zebrafish studies and help address some of the existing challenges to accelerate progress in the use of zebrafish to understand neurodevelopmental disorders, and ASD specifically, will be further discussed in the coming months.
References
- Ahrens M.B. et al. Nat. Methods 10, 413–420 (2013) PubMed
- Anderson J.L. et al. PLoS Genet. 13, e1007105 (2017) PubMed
- Pardo-Martin C. et al. Nat. Methods 7, 634-636 (2010) PubMed
- Rennekamp A.J. and Peterson R.T. Curr. Opin. Chem. Biol. 24, 58-70 (2015) PubMed
- Rennekamp A.J. et al. Nat. Chem. Biol. 12, 552-558 (2016) PubMed
- Hoffman E.J. et al. Neuron 89, 725-733 (2016) PubMed
- Eimon P.M. et al. Nat. Commun. 9, 219 (2018) PubMed
- Report of the National Advisory Mental Health Council Workgroup on Genomics: Opportunities and Challenges of Psychiatric Genetics (2018) Article
- Marquat G.D. et al. Gigascience 6, 1-15 (2017) PubMed
- Gupta T. et al. Methods Epub ahead of print (2018) PubMed
- Farrell A.J. et al. Science 360, eaar 3131 (2018) PubMed
- Pandey S. et al. Curr. Biol. 7, 1052-1065 (2018) PubMed
- Raj B. et al. Nat. Biotechnol. 36, 442-450 (2018) PubMed
- Ijaz S. and Hoffman E.J. J. Am. Acad. Child. Adolesc. Psychiatry 55, 746-748 (2016) PubMed