
SFARI Gene is a resource provided by the Simons Foundation Autism Research Initiative (SFARI) that allows researchers to keep track of the ever-expanding list of autism genetic risk factors through access to a publicly available, searchable and evolving online database. SFARI Gene tracks and integrates a variety of genetic data generated by research studies, including data from linkage and association studies, cytogenetic abnormalities, specific mutational analyses and relevant animal models. Each autism risk gene is carefully evaluated and classified using a set of annotation rules initially developed by the SFARI Gene Advisory Board and subsequently modified as new data emerge.
The SFARI Gene database was developed under the name AutDB by MindSpec Inc., a nonprofit organization that utilizes bioinformatics strategies to build disease-specific databases in order to accelerate research on neurodevelopmental disorders. In 2008, MindSpec Inc. licensed the database to SFARI, and through a collaborative effort with SFARI, MindSpec Inc. scientists continue to maintain and update the database on a quarterly basis.
The SFARI Gene website recently underwent a relaunch. I spoke with Sharmila Banerjee-Basu, founder and chief scientific officer of MindSpec Inc., to discuss how SFARI Gene has evolved over the years. The interview has been edited for clarity and brevity.
What was the rationale behind the establishment of the autism database that is now known as SFARI Gene?
I have both a professional and personal interest in the genetic architecture underlying autism. The idea of this project first came up in 2006, when I realized that there weren’t any resources to help researchers if one was looking for a set of genes associated with autism. With the help of two IT professionals, Ravi Kollu and Saumyendra N. Basu, I decided to build a resource that would select for autism genes — that is genes that are associated with the risk of having the disorder — in an unbiased fashion. One of my earliest goals, which is still valid now, was to make this an open-access resource so that any researcher who is interested in doing any kind of work on autism could have access to a curated resource of autism genetics.
“Our goal is to make [SFARI Gene] a cutting-edge resource and to make sure it is current and dynamic.”
SFARI Gene is regularly updated to take into account new scientific research. How does MindSpec Inc. track and evaluate new autism genetics data?
When the data was first released, I was the only one who was annotating it, and now we have a group of really talented scientists working together on this. We track all genetics papers that get published, and we take that data and use our scientific-annotation model and put it into the database. Our goal is to make this a cutting-edge resource and to make sure it is current and dynamic. With our first release, the number of autism-associated genes was less than a hundred. With the most recent release, we just approached 900 genes. We are constantly scanning the literature, evaluating the papers and curating the database.
SFARI Gene contains data from studies of both rare and common genetic variants. How does SFARI Gene handle these distinct data sets?
In the initial model that was published in 20091, I postulated an integrated model of rare and common variants. At that time, there were very few rare variants. That has really expanded in the last five or six years, due to studies taking advantage of the Simons Simplex Collection and other cohorts like it.
The common variants were also not very robust back then, since most of the genetic association studies were underpowered. In fact, we still do not have good genome-wide association studies (GWAS) for common variants. We do not have the numbers like they do in the schizophrenia field, where the GWAS studies have been able to identify good common variants. And we are waiting for those high-powered GWAS studies to come out, so that we will have common variants that have genome-wide significance.
For rare variants, coming from whole-exome sequencing studies, there are many ways of looking at their frequency distributions and seeing how they compare to the typical control population. But for common variants, a different set of statistical tests is used. So when we annotate common and rare variants, we keep them in two different categories and we consider them in a separate manner.
SFARI Gene contains a number of modules. Could you talk briefly about this modular setup?
The first module that went public and that was made openly available was the Human Gene module. When we released the resource, I got an invitation from the science team at SFARI to come and present my project — that was in 2007. The result of this was a collaboration with SFARI and support to create SFARI Gene2.
Our overall goal is to get a systems view of autism since the genetic architecture is highly heterogeneous. There are so many genes. With that in mind, the project started expanding, and with generous funding from SFARI, we added additional modules.
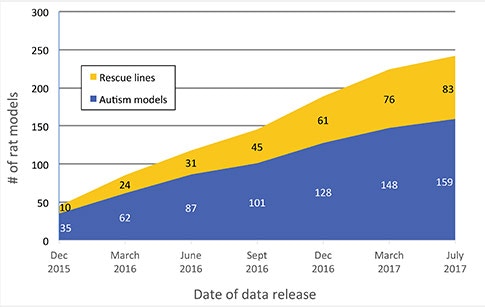
Two of the modules that were added were the Animal Models module and the Protein Interaction Network (PIN) module. The Human Gene module gives the genetic architecture, but the Animal Models and PIN modules are giving deeper insights into the function of these genes.
Another module, the Copy Number Variant (CNV) module, is also very highly accessed. The Human Gene module concentrates on a single gene at a time. But there are multigenic CNVs that are associated with autism. And there are many research groups that use our CNV data, like the Simons Variation in Individuals Project (Simons VIP), which studies individuals with CNVs in 16p11.2 or 1q21.1. So that’s also another module that is an important player.
The Animal Models module contains information from mouse and rat models. Are there plans to expand this to include additional animal models?
We are working actively to look into other species like Drosophila and zebrafish, as that literature is catching up. There are many things that can be done in a much faster way in these model organisms, and, of course, there are also other advantages of using these models compared to rodents. We are curating those datasets now, and hopefully by the end of the year, we will have a release of the new species.
But if you look into different areas of publications, the mouse models research is probably where we are getting the deepest insights into biology of autism. At present we are working hard to annotate those models to include more brain regions, neuronal types, circuits and electrophysiological findings.
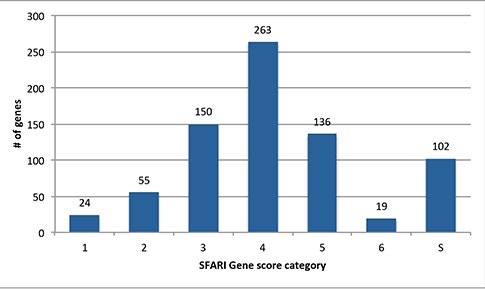
A very popular service that SFARI Gene provides to the research community is the Gene Scoring module. This module evaluates the data in SFARI Gene, providing a ranking for each autism risk gene in the database based on an assessment of the strength of human genetic evidence. How is the gene scoring done and how are confidence values assigned?
So this is a really good question. In fact, I just came back from a meeting and there were so many talks where they showed the SFARI Gene score for their gene of interest. If somebody’s gene falls into the high-confidence category [in the Gene Scoring module], they get really excited.
When we started this project, it was a collection of all the different types of genetic evidence that are associated with autism. In order to evaluate the information, Alan Packer (SFARI senior scientist) and I created an Advisory Board consisting of six scientists. Together, we came up with the gene-scoring criteria on how to evaluate different types of scientific evidence. Although the scoring criteria on the SFARI Gene website is only one page long, it took more than a year to finalize it2, with many meetings and teleconferences and so on. But that scoring criteria is also quite dynamic. We continue to maintain the database, and it’s really a cutting-edge resource.
Back in 2006 when this project started, I wanted to make sure the database stayed current. There are so many academic resources resulting from the publication of really good papers, but then the person responsible maybe moves on to a different project and the database doesn’t get updated. There are so many examples of that. That was something in my very first meeting with SFARI that I mentioned. I said that our goal had to be to keep the resource cutting-edge but also very dynamic. And I must say that I have a lot of regard for the SFARI team. As new evidence comes, we all work together and always discuss the work. We keep updating the set of scoring rules and, with each gene that is discovered, we put it through those filters, put a score to it, and then the gene goes into the database for everybody to use.
How is the SFARI Gene database changing the way researchers think about, study and evaluate autism?
When we started this project 10 years ago, our understanding of the genetic architecture of autism was really in its infancy. Now we have more inclusive, comprehensive models and a better understanding of the pathways involved. Researchers can assess gene patterns, like differential expression in a brain region, tissue or under certain physiological conditions and relate that to autism phenotypes. A paper came out earlier this year where researchers used the database to make certain links between glutamatergic neurotransmission and autism pathology3. Because of the comprehensive architecture of our database, they found important connections to autism genes that they wouldn’t have found in whole-exome datasets alone.
Also, with all of the work on autism and related neurodevelopmental disorders, we are getting a broader picture of the genetic basis of human cognition. We are learning more about how human cognition is shaped by genetics. And there is still lots to do. This resource has evolved over the last 10 years. My anticipation is it is going to evolve more, and it will keep up with our expanding knowledge of the genetic architecture of autism.
As you say, since the initial launch of the SFARI Gene platform in 2008, it has expanded in numerous ways to meet the needs of the scientific community. In the coming years, what are your dreams for the next evolution of SFARI Gene?
I can tell you exactly. It’s the human phenotypes. Lots of genetic variants are now being identified in individuals with autism, but from a phenotype perspective, autism has a very broad clinical presentation and not every symptom is present in every child or adult. Also, the behavioral diagnostic criteria are limited to social and repetitive behaviors, whereas, the autism profile is much broader.
One of the things clearly in every researcher’s mind is how you can link genotypes to autism phenotypes. So I would really like to annotate the phenotypes, the definitive features, associated with genetic subtypes. That has a lot of implications for the treatment and management of the disorder. My hope and my goal is that, one day, our database will help advance definitive testing for autism.


