
In 2020, the Simons Foundation Autism Research Initiative (SFARI) spearheaded an effort to develop a roadmap that could address fundamental questions on the interplay between the microbiome and the brain in autism spectrum disorder (ASD). SFARI consultant-in-residence Gaspar Taroncher-Oldenburg has been leading this project, with the goal of elucidating the potential role of the microbiome in ASD and identifying areas of research that SFARI could help advance on this topic. I recently spoke with Taroncher-Oldenburg to learn more about the state of knowledge in the field, his collaboration with SFARI and his views on how this work could increase our understanding of a potential causal connection between the microbiome and ASD.
Taroncher-Oldenburg is a microbiologist and translational scientist with extensive experience in establishing and leading multidisciplinary teams to advance innovation in complex areas such as the microbiome and disease.
The interview has been edited for clarity and brevity.
Why look at the microbiome in ASD now?
It is hardly a secret that over the past two decades, the central role the microbiome plays in human biology, and specifically in disease, has come into greater focus. If you do a simple literature search for “human microbiome,” you will find that the number of publications in the field started rising dramatically around 2010. This is a direct consequence of the scientific community’s gradual recognition that trillions of microbes colonize nearly all sites on our bodies, and that these microbes — collectively known as the microbiome — are instrumental in determining and regulating our physiology. Equally important, access to unparalleled and rapidly evolving sequencing capabilities brought on by the human genome project has enabled the massive sequencing of the metagenomes associated with the microbiomes. Not surprisingly, as the role of the microbiome in human biology was being firmly established, determining its potential contribution to disease followed. Today, there is an impressive and expanding body of recognized associations between the microbiome and human diseases, including ASD.
If I understand correctly, most of the evidence we have up to this point provides a correlational association between ASD and the microbiome. What types of evidence do we have, and why do you think it warrants pursuing efforts to establish a causal link?
You are correct that most of our evidence to date is correlational. Metagenomic analyses of the human gut microbiome in ASD have revealed many, albeit inconsistent, variations in microbial diversity in individuals with ASD compared with neurotypicals.1-3 Metagenome-based functional reconstructions and metabolic analyses have also shown strong, but still inconclusive, differences between individuals with and without ASD.4-6 Observational, cross-sectional, human studies provide further correlational links. For example, the density of the mucin-degrading bacterium Akkermansia muciniphila, a bacterium generally considered to be beneficial in the gut, has been linked to the severity of ASD symptoms.7 But there are a few highly publicized instances in which direct or indirect microbiome manipulations have resulted in positive outcomes for individuals with ASD and their families.8
“These studies give us reason to believe that microbiome manipulations may have a causal role, and clinical value, in treating ASD features. But solving outstanding questions around microbiome composition, and potential microbial drivers of ASD, will require further well-controlled and well-powered human studies that integrate different omics level analyses.”
Probably the most complete longitudinal intervention study to date is an open-label fecal microbiota transplantation (FMT) study carried out at Arizona State University from Kang et al.9 assessing eighteen children with ASD. Two months after a FMT from neurotypical donors, the children with ASD exhibited significant improvements in gastrointestinal symptoms measured through the Gastrointestinal Symptom Rating Scale (GSRS) and daily stool records (DSR), as well as behavioral core features of ASD, which were assessed through a number of questionnaires and interviews, including the Parent Global Impressions-III (PGI-III), the Childhood Autism Rating Scale (CARS), the Social Responsiveness Scale (SRS) and the Vineland Adaptive Behavior Scale II (VABS-II). A shift in plasma metabolites toward profiles more similar to those of neurotypical children was also observed. These improvements were accompanied by significant changes at the microbiome level, including an increase in bacterial diversity to levels more in line with those observed in neurotypical children and an enrichment in beneficial bacteria such as Bifidobacteria and Prevotella. Even more encouraging is the fact that a two-year follow up of the entire cohort revealed further improvement in behavioral symptoms, with the gastrointestinal symptom profiles and shifts in microbiome composition observed at the two-month mark also maintained.10
These studies give us reason to believe that microbiome manipulations may have a causal role, and clinical value, in treating ASD features. But solving outstanding questions around microbiome composition, and potential microbial drivers of ASD, will require further well-controlled and well-powered human studies that integrate different omics level analyses.
Could you elaborate more on the sorts of human studies that you think are needed to help uncover potential causal links between the microbiome and the brain?
It’s a complex task and, as exciting as the results from existing interventional studies are, they also lay bare some critical shortcomings that need to be addressed. There are ongoing controversies about how you even measure and quantify the microbiome — various technical and statistical biases make it difficult to compare across microbiome studies. You have many of the same issues with ASD — for example, bias toward males because ASD presents mostly in boys and biases towards cohorts on the less severe end of the spectrum because they are easier to work with. To address this, we need better powered and controlled longitudinal studies, and more robust and objective behavioral and physiological measures for quantifying and evaluating treatment outcomes. We also need an integrated data analysis approach involving multi-omics sampling to capture the complex genetic, metabolic, neuroimmune, neuronal and neuroendocrine networks that constitute the microbiome-gut-brain axis. Making this happen hinges on two things: one, bringing together an engaged and multidisciplinary community and two, providing a framework focused on an output that will be more than the sum of its parts.
“We needed to do something different that takes into account all of the complexity of the microbiome and ASD.”
You have been leading SFARI’s effort to elucidate the potential link between the microbiome and ASD. What approach are you taking to address the existing challenges and help to move the field forward?
One of the first steps we took was to engage, during the second half of 2020, with close to fifty researchers in the fields of microbiology, neurology, immunology, computation and computational biology, and clinicians who were interested in the microbiome–gut–brain–ASD axis. We solicited their thoughts on the SFARI microbiome-in-ASD initiative, and the response was overwhelmingly positive. The view was that it’s reasonable to think that there might be something going on, but we need to learn a lot more about how the microbiome and ASD might be interacting.
One of the most consistent points of feedback we got was that we have to think about this question on two levels. If we want to learn something about causal mechanisms, we should think about long-term observational studies looking very early, even perinatally, and at different times in development. But a more immediate goal should be to design better intervention studies to allow insights on the clinical side. It’s important to remember that, for families, a 50 percent improvement in ASD symptoms is a world of difference. But the type of improvement might be different for every individual. So we needed to do something different that takes into account all of the complexity of the microbiome and ASD.
What did “doing something different” mean to you?
We decided that a good starting point would be to do a meta-analysis of the existing data. Not a regular meta-analysis, but a different kind of meta-analysis that would aim to extract previously untapped information contained in those same studies. The vast majority of existing studies have been assessed using traditional statistics to determine compositional changes using group averages. The way that works in ASD microbiome studies is that you take the average counts for an ASD cohort and compare it to the average of a neurotypical cohort. This averaging approach means that you actually significantly dampen many, if not most, biological signals in your data because you dilute any signal associated with variables such as age or sex. But there’s another type of statistics, called Bayesian statistics, that allows you to evaluate the data across cohorts on an individualized basis to maximize your comparative power. We chose to match the data according to age and sex because of the relevance of these two variables when it comes to microbiome development and ASD phenotype, respectively (Figure 1).
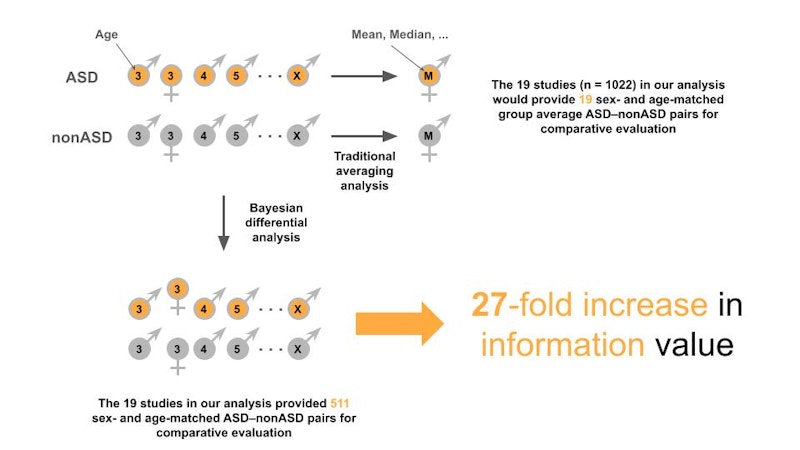
Next, we focused on determining microbial log ratios, a measure of perturbation, rather than absolute microbiome compositional changes to overcome batch and other experimental confounders inherent to the microbiome, and by extension, most omics datasets. Our hope was that such a Bayesian-based “mega-analysis” approach on the existing cohort studies would allow us to detect real changes in microbial dynamics correlated with ASD.
The idea was very simple — develop a framework for doing integrated, multi-omic analyses that would allow us to coordinate information across multiple levels of biological organization and information in the existing ASD microbiome studies. The process itself, however, was complex — it needed computational know-how and access to sufficient computational power. Lucky for us, we were able to join efforts with Jamie Morton in Richard Bonneau’s group at the Center for Computational Biology of the Flatiron Institute, a research division of the Simons Foundation focusing on advancing scientific research using a variety of computational methods. A major focus of this collaboration was to develop an effective multi-omic analysis approach that would allow us to coordinate data across multiple levels of biological organization and information, preempting the natural tendency to address questions by drilling down in great detail at each omic level. The hope was that such an approach might reveal emergent properties that were not well captured by existing data analysis techniques.
“The analysis approach we’ve taken is very powerful because of its simplicity. We’re at the point now of thinking about how we translate this to inform future work so that we can start pulling out really interesting and valuable functional information.”
What are some of the interesting findings that have come out of your ongoing collaboration with the Flatiron Institute?
Getting the analysis pipeline in place was complex and took a while. But now that we have it in place, we have been amazed. We’ve never seen such an overlap in microbiome studies in any meta-analysis before. The data that we have, which we’re in the process of submitting for publication, show very high levels of signal consistency across studies within individual omics levels, and this remarkable convergence across different levels of omics information in ASD.
And we had the data from the Kang et al. FMT intervention study to use for validation of what we are seeing. That analysis showed us that, when you go from pre-FMT treatment to post-FMT treatment in children with ASD, the post-FMT data move toward the neurotypical group. We performed a number of other analyses and comparisons as well. In particular, an evaluation of a Pitt-Hopkins syndrome cohort in the context of our overall analysis showed the potential power of stratifying any future studies by underlying ASD genotype. Of course, these data still reflect hundreds of metabolites and genes, but they tell us that there are strong correlations between microbiome composition and ASD. It’s just a very complex picture, which is not a surprise since the microbiome is complex and so is ASD.
What do you see as the next steps for SFARI to help drive future work in understanding the microbiome in ASD?
The analysis approach we’ve taken is very powerful because of its simplicity. We’re at the point now of thinking about how we translate this to inform future work so that we can start pulling out really interesting and valuable functional information.
Moving forward, our data suggest that interventional and longitudinal studies are probably the best way to go, and with a focus on microbiome functionality. SFARI could help streamline access to potential study participants through SPARK and the recent research matching program to help seed these types of integrated, long-term studies. Unifying the genetic background as much as possible will also help — a specific cohort, a specific gender or a specific behavior or behavioral spectrum. We also need to work on getting precise measures of the metabolome at the serum level and at the urine level and, if it can be standardized, at the fecal level in such cohorts. If we design intervention studies properly, we could infer something about the underlying biology as well. SFARI is in an excellent position to convene and organize a multidisciplinary community of researchers to help drive such studies.
What do you think the ultimate impact will be on the community, especially on individuals with autism and their families?
We obviously don’t know what the outcome of all this will be, but we can say one thing for sure: this work will give us a much better grasp of the role — big or small — of the microbiome in ASD. We are looking at this complex system from many angles, including time frames relevant to therapeutic and even preventative intervention. In the end, we are aiming to gain a better understanding of potential mechanisms underlying ASD while also advancing real solutions for individuals with ASD and their families.
More information about this study can be found in Morton et al., bioRxiv, 2022. Updated 3/10/2022
References
- Andreo-Martínez P. et al. J. Autism Dev. Disord. Online ahead of print (2021) PubMed
- Bezawada N. et al. Ann. Nutr. Metab. 76, 16-29 (2020) PubMed
- Xu M. et al. Front. Psychiatry 10, 473 (2019) PubMed
- Likhitweerawong N. et al. Metab. Brain Dis. 36, 1641-1671 (2021) PubMed
- Zhu J. et al. J. Autism Dev. Disord. Online ahead of print (2021) PubMed
- Xu X.-J. et al. Front. Psychiatry 12, 657105 (2021) PubMed
- Wang L. et al. Appl. Environ. Microbiol. 77, 6718-6721 (2011) PubMed
- Svoboda E. Nature 577, S14-S15 (2020) PubMed
- Kang D.-W. et al. Microbiome 5, 10 (2017) PubMed
- Kang D.-W. et al. Sci. Reports 9, 5821 (2019) PubMed


