Research into the genetics of autism spectrum disorder (ASD), including work supported by the SFARI initiatives, has resulted in tremendous progress into our understanding of the complex genetic makeup of ASD. Hundreds of genes have been associated with increased risk of ASD, but this wealth of knowledge presents its own set of challenges. In particular, our ability to identify such changes greatly surpasses our understanding of how these genetic changes affect neuronal function and lead to the condition.
A recent study, supported in part by an Explorer grant to SFARI Investigator Robi Mitra, presents a cellular system that allows one to explore the function of high-confidence ASD risk genes in neurodevelopment — using a scalable functional genomics approach.
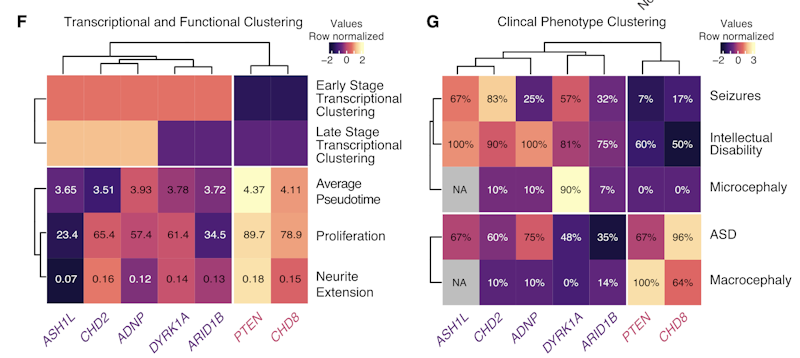
Mitra and colleagues used CRISPR/Cas9 technology to transcriptionally repress high-confidence ASD risk genes. They then examined the effects on transcriptional profiles and neuronal development using single-cell RNA sequencing of differentiating LUHMES neuronal progenitor cells. The LUHMES cell line has been used previously to model neurodevelopmental disorders (Shah et al. Wellcome Open Res. 2016; Matelski et al. Mol. Psychiatry 2020). The researchers then assessed how reductions in 13 different high-confidence ASD risk genes affected the neurodevelopmental timeline, demonstrating that four of the genes (CHD2, ASH1L, ARID1B and DYRK1A) caused a delay in neuronal differentiation while two (PTEN and CHD8) accelerated the process.
Knockdown of these genes also affected larger transcriptional networks. Clustering transcriptional changes by their effects on the differentiation timeline and their gene ontology suggested that alterations in these genes disrupt core functional networks. This also led to the identification of ADNP as an additional risk gene that delays neuronal differentiation.
Mitra’s team found that the early “delay” effects appear to reflect a disruption of the G2/M cell cycle transition and negatively regulate early cell development, while later delays reflect defects in neuronal maturation steps. In contrast, PTEN and CHD8 knockdown-induced acceleration of differentiation did not appear to reflect a common pathway. Their findings also fit with recent work on affected individuals, with alterations in CHD2, ARID1B, DYRK1A and ADNP being associated with more severe neurodevelopmental delays, while changes in CHD8 and PTEN were not (Stessman et al. Nat. Genet. 2017).
It’s especially noteworthy that Mitra and colleagues found that their functional networks mirrored an overlap in clinical phenotypes. Individuals with mutations in the “delay” ASD risk genes were more likely to present with intellectual disability and microcephaly, while those with the “accelerate” risk genes were more likely to have macrocephaly.
The findings from this small sample of ASD risk genes represent an important step forward for the field. Further expansion of this scalable and cost-effective system will enable greater insights into convergent functional and clinical phenotypes and will improve our mechanistic understanding of neurodevelopmental disorders.
Reference(s)
High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation.
Lalli M.A., Avey D., Dougherty J., Milbrandt J., Mitra R.


