Missense mutations make a substantial contribution to autism risk, as shown by numerous DNA sequencing studies. The SFARI Functional Screen of Autism-Associated Variants request for applications (RFA) sponsors the development of methods and platforms to identify those mutations that are truly pathogenic. A new study outlined the use of multiple model organisms to exhaustively characterize the functional impact of many missense mutations identified in a single gene, and in so doing established a pipeline for additional ASD risk genes to be studied in a similar manner.
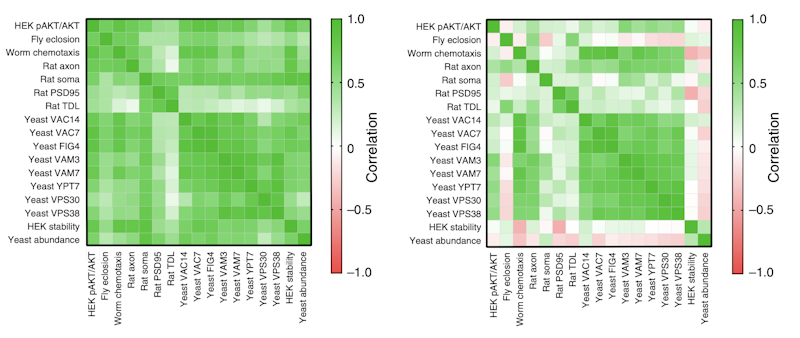
This work was supported by a targeted award under the ‘Functional Screen’ RFA to SFARI Investigator Kurt Haas, who led a mini-consortium of researchers working on the ASD risk gene PTEN. Haas and colleagues examined 106 missense and nonsense mutations in 18 assays from five model systems, including yeast, Caenorhabditis elegans, Drosophila melanogaster, primary rat hippocampal neurons and human cell lines. These assays allowed them to test for functional impact in a range of ways, including effects on protein stability (yeast and human cells), development (Drosophila), neuronal morphology (rat) and chemotaxis (C. elegans), among others. The variety of functional data enabled the researchers to reach several conclusions. First, there was strong, if not perfect, correlation of effects among the functional assays for each variant. Second, a significant number of variants showed dominant negative effects. Third, a substantial majority of the variants had effects on protein stability, with those whose impact was more independent of effects on stability showing weaker cross-model correlation. Finally, they were able to use the data to make a prediction of relative pathogenicity for each variant.
Haas and colleagues have conducted similar experiments on missense variants in SYNGAP1 (Meili et al., bioRxiv, 2020) and have an additional SFARI award to examine other ASD risk genes. Ultimately, one might expect these results to be incorporated into the American College of Medical Genetics guidelines (Richards et al., Genet. Med., 2015) for assessing pathogenicity of mutations.
Reference(s)
Multi-model functionalization of disease-associated PTEN missense mutations identifies multiple molecular mechanisms underlying protein dysfunction.
Post K.L., Belmadani M., Ganguly P., Meili F., Dingwall R., McDiarmid T.A., Meyers W.M., Herrington C., Young B.P., Callaghan D.B., Rogic S., Edwards M., Niciforovic A., Cau A., Rankin C., O'Connor T., Bamji S., Loewen C. J., Allan D., Pavlidis P., Haas K.


