Mutations in genes encoding a number of chromatin-associated proteins are associated with Kleefstra syndrome spectrum (KSS), a neurodevelopmental condition typically characterized by intellectual disability and autism spectrum disorder (ASD). This convergence on a similar phenotype suggests that these mutations must have common biological consequences on brain function despite the distinct biochemical properties of the gene products. A new study sheds light on the nature of this biological convergence on neuronal activity.
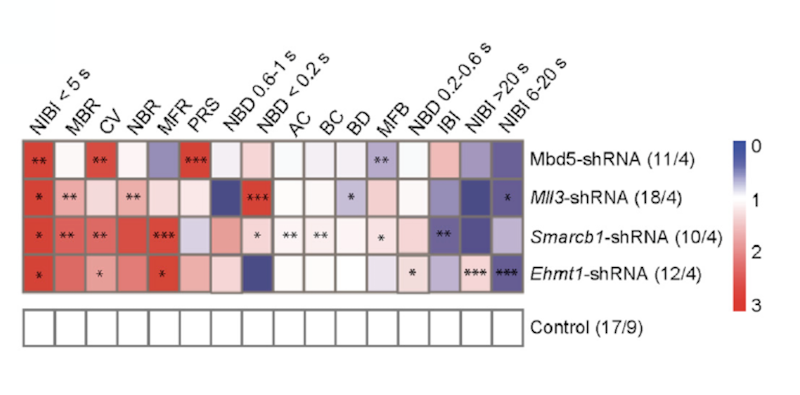
The work was supported in part by a Pilot Award to SFARI Investigator Nael Nadif Kasri. The authors examined the downstream effects of mutations in four KSS-associated genes: EHMT1, MBD5, MLL3 and SMARCB1. Using micro-electrode arrays, the researchers were able to show that knockdown of EHMT1, MLL3 or SMARCB1 expression in rat cortical cultures each resulted in a higher level of neuronal network activity (network burst rate). Knockdown of each of the four genes showed impairments in the pattern of network burst activity late in development. Kasri and colleagues also showed that knockdown of each gene was associated with changes in intrinsic properties that directly or indirectly affect the generation of action potentials, leading in each case to different levels of increased neuronal excitability. Experiments focused on neuronal excitatory-inhibitory balance revealed that the ratio of excitation and inhibition was shifted to increased excitation due to reduced inhibitory synaptic input.
Transcriptomic analysis showed that EHMT1-, SMARCB1- and MLL3-deficient networks had high similarity in the genes whose expression was altered. These included 34 common genes that had previously been linked to the range of phenotypes found in KSS. Interestingly, the direction of change of many of these genes was not always the same, lending support to the idea that the common clinical phenotypes likely arise from convergence at the level of the neuronal network.
Reference(s)
Distinct pathogenic genes causing intellectual disability and autism exhibit a common neuronal network hyperactivity phenotype.
Frega M., Selten M., Mossink B., Keller J.M., Linda K., Moerschen R., Qu J., Koerner P., Jansen S., Oudakker A., Kleefstra T., van Bokhoven H., Zhou H., Schubert D., Kasri N. N.


