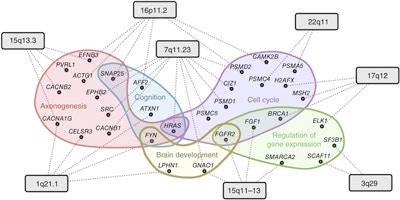
Although sequencing studies have identified more than 60 candidate autism risk genes over the last several years, a significant number of risk genes remain to be discovered. Olga Troyanskaya (a Simons Center for Data Analysis scientist) and her colleagues developed a machine-learning approach that utilizes a brain-specific gene functional interaction network to predict autism risk genes across the genome. The tool identified hundreds of autism risk genes, including many previously unreported candidate genes, along with key cellular pathways and developmental stages of the brain that are potentially dysregulated in autism. Many of the candidate genes have since been validated by independent studies — including Simons Simplex Collection exome-sequencing studies — as genuine autism-associated genes. The researchers also used their tool to prioritize risk genes within eight of the most common autism-associated copy number variants (CNVs). This computational approach is expected to help advance gene discovery efforts by prioritizing genes for focused experimental studies. All of the predictions and data from this study are accessible via a dedicated web portal.
Reference(s)
Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder.
Krishnan A., Zhang R., Yao V., Theesfeld C.L., Wong A.K., Tadych A., Volfovsky N., Packer A., Troyanskaya O.G.


