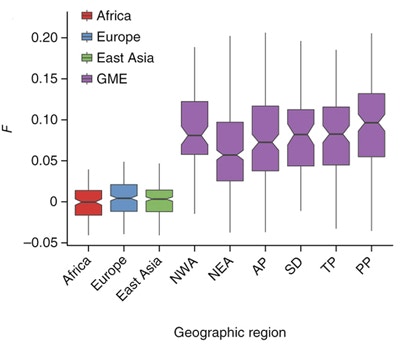
The Greater Middle East (GME) population is known to show an elevated burden of recessive Mendelian disease, due to the tradition of consanguinity that is practiced in a number of countries in this region. To better understand the extent of rare genetic variation in this population, SFARI Investigator Joseph Gleeson and his colleagues in the GME Variome Consortium generated a whole-exome variome from 1,111 unrelated individuals from GME regions. The researchers performed a number of initial characterizations of the dataset, including a pilot study of recessive hereditary spastic paraplegia, to illustrate its application. The GME variome is a publicly accessible resource and its use in future sequencing projects is expected to aid in the identification of causative recessive genes linked to diseases of all classes, including autism spectrum disorder.
Reference(s)
Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery.
Scott E.M., Halees A., Itan Y., Spencer E.G., He Y., Azab M.A., Gabriel S.B., Belkadi A., Boisson B., Abel L., Clark A.G., Greater Middle East Variome C., Alkuraya F.S., Casanova J.L., Gleeson J.


