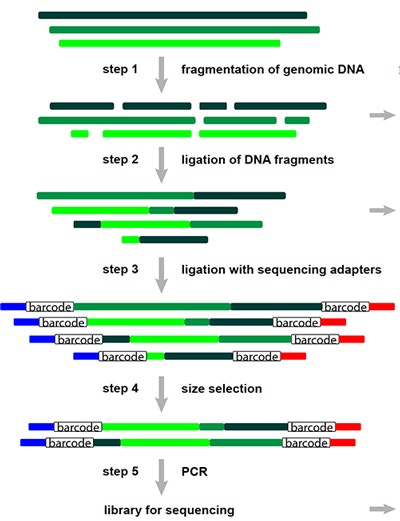
Analyses of copy number variants (CNVs) have contributed significantly to our understanding of the genetic etiology of a variety of diseases, including autism spectrum disorder (ASD). Despite this, a large portion of ASD risk remains unexplained. Detection of CNVs has been limited, in part due to the significant costs associated with both chromosomal microarray analyses (CMA) and whole-genome sequencing (WGS). SFARI Investigator Michael Wigler and his colleague Dan Levy at Cold Spring Harbor Laboratory in New York have developed a new fragmentation and sequencing method for CNV analysis called SMASH (Short Multiply Aggregated Sequence Homologies). Using data from the Simons Simplex Collection (SSC), the researchers have demonstrated that this methodology generates high-resolution CNV data of comparable quality to that obtained by WGS analyses, but at a lower cost. The SMASH protocol will open up new avenues for assessing genetic risk in ASD.
Reference(s)
SMASH, a fragmentation and sequencing method for genomic copy number analysis.
Wang Z., Andrews P., Kendall J., Ma B., Hakker I., Rodgers L., Ronemus M., Wigler M., Levy D.


