Frameshift
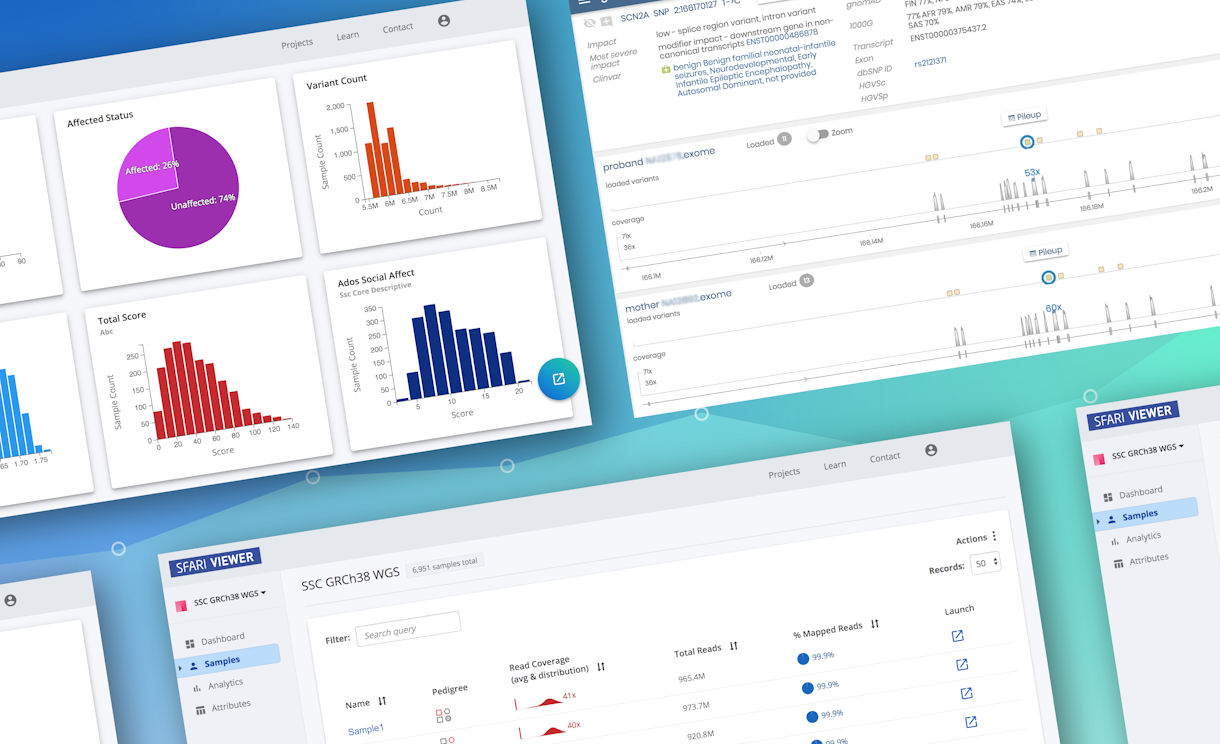
SFARI Viewer, a new online tool that enables researchers to visualize, explore and analyze SFARI genomic and phenotypic data in real-time, has launched. This tool was developed by Frameshift, Gabor Marth’s team at the University of Utah and the SFARI Informatics group led by SFARI director of data and analytics Natalia Volfovsky.
The platform currently hosts whole-exome and whole-genome sequencing data from the Simons Simplex Collection (SSC) and whole-exome sequencing data from SPARK (Simons Foundation Powering Autism Research for Knowledge), in addition to associated phenotypic information for these cohorts (Table 1).
Table 1. Genetic data available in SFARI Viewer. Data include whole-exome and whole-genome sequencing data for over 9,000 individuals (including 2,406 individuals with ASD) from the Simons Simplex Collection (SSC) as well as the first data release from SPARK (Simons Foundation Powering Autism Research for Knowledge), comprising 1,379 exomes (including 472 individuals with ASD). Additional genomic data will be added to the platform in the coming months. SSC genomics data were aligned to either the Genome Reference Consortium human genome build 37 (GRCh37) or GRCh38.
| SFARI dataset | # of samples1 |
|---|---|
| SSC GRCh37 | 9,046 exomes |
| 2,135 genomes | |
| SSC GRCh38 | 9,078 genomes |
| SPARK Pilot | 1,379 exomes |
1Note: The number of samples available in the platform is currently slightly lower than the numbers shown here, due to ongoing migration of data files from SFARI Base. All sequence files are expected to be available within the next few weeks.
SFARI Viewer adds to the body of data-analysis tools that SFARI has supported in recent years to help researchers navigate through the genetic and phenotypic data acquired from the different SFARI cohorts. These tools include the WuXi NextCODE SSC portal, a cloud-based database that allows for the visualization and analysis of whole-exome sequencing data and associated phenotypic data from the SSC, and the Genotypes and Phenotypes in Families (GPF) tool — an online platform developed by SFARI Investigator Ivan Iossifov that enables users to analyze whole-exome and whole-genome sequencing data, as well as single nucleotide polymorphism genotyping data and associated phenotypic information from the SSC, SPARK and the Simons Variation in Individuals Project (Simons VIP).
What distinguishes SFARI Viewer from existing SFARI analytic tools is that researchers are able to dynamically interact with the data in real time. Users can filter the data sets by selecting portions of the data (e.g., only samples with both nonverbal and verbal intelligence quotient [IQ] scores greater than 110) and create custom projects based on the investigators’ particular research questions. This feature may help investigators to replicate previous findings by re-creating the samples analyzed in other studies, as well to select samples from SFARI data sets that are comprised of individuals similar to those in a researcher’s own cohort. Furthermore, users can compare phenotypic data in custom-created projects with all SFARI data sets and visually compare data from different subsets to search for potential correlations between phenotypes. Crucially, these operations are built upon an intuitive interface, so that no prior computational or bioinformatics experience is required to perform these tasks (Video 1).
By clicking to watch this video, you agree to our privacy policy.
In addition to using these built-in functionalities, researchers can perform specific tasks and further probe the data by launching additional web-based applications directly from SFARI Viewer. One of these applications, gene.iobio, annotates variants in a user-defined gene list for any of the SFARI families, thus making it possible to identify potentially interesting variants in the proband based on characteristics, such as population allele frequency or the presence of that variant in the ClinVar database.
Alistair Ward, president and COO of Frameshift, had presented an earlier prototype of SFARI Viewer at a workshop on SFARI Genomics platforms that was held at the Simons Foundation last spring. “We are excited to see the live version of the platform finally being released and look forward to learning about the creative ways researchers will use it,” he says.
Adds Volfovsky, “We tried to make SFARI Viewer as interactive and user friendly as possible. We look forward to receiving feedback from investigators, so we can continue to improve the functionalities of this tool to best serve the needs of the autism genetics research community.”
More information, including ‘Tutorials’ and ‘Get Started’ pages on how to use SFARI Viewer, can be found here. The portal also offers an instant-chat tool for users to ask questions directly. For those who may require a more formal introduction to the tool, the developers are available to provide step-by-step tutorials of the platform and answer questions via conference calls with individual groups.
To send your feedback, please contact: [email protected]


