Since launching the Simons Simplex Collection (SSC)1, the Simons Foundation Autism Research Initiative (SFARI) has supported a number of other initiatives whose aims include collecting and sharing large-scale genotypic and phenotypic data from individuals with autism and related neurodevelopmental disorders [e.g., the Simons Variation in Individuals Project (Simons VIP)2 and Simons Foundation Powering Autism Research for Knowledge (SPARK)3]. As the amount of information generated by these projects keeps growing and becoming more complex, SFARI is now investing in analytical tools that can efficiently manage and integrate all datasets, while providing easy data access and exploration for users in the research community.
Priorities of this effort include harmonizing the data, which are distributed across different projects and located in different archives (e.g., Amazon Web Services, local servers), implementing options that enable users to deploy their own analytical tools to explore SFARI datasets and ensuring that participants’ data are shared securely. To support international collaborations, governmental restrictions for biomedical data usage and sharing across jurisdictions also need to be considered.
To begin addressing these challenges, SFARI held two workshops in the fall of 2016 that focused on (1) optimizing data storage and sharing environments, and (2) creating web interfaces to enable a wide range of researchers to visualize and analyze genomics data. Speakers at these workshops discussed examples of computational and data-sharing environments that are successfully used in fields outside neuroscience, such as cancer biology, astronomy and particle physics.
On April 10, 2018, SFARI organized a third workshop that presented two web interfaces that facilitate the exploration and visualization of SFARI genomics data: the Genotypes and Phenotypes in Families (GPF) platform and Hub.iobio. A summary of these presentations is included below.
Genotypes and Phenotypes in Families (GPF) platform
Developed by SFARI Investigator Ivan Iossifov and his team at Cold Spring Harbor Laboratory and the New York Genome Center, the GPF platform helps users visualize and analyze genetic and phenotypic data from SFARI collections (SSC, Simons VIP and SPARK). Genetic data include genotyping array data and whole-exome and whole-genome (where applicable) sequencing data.
Six distinct datasets are currently available on this platform (Table 1). The ‘Sequencing de novo dataset,’ which comprises de novo variants associated with autism (based on data from the SSC) and other neurodevelopmental disorders (i.e., congenital heart disease, intellectual disability, epilepsy, schizophrenia), was assembled by Iossifov and his team from the published literature and is publicly available. Users must request access (via a SFARI Base application) to analyze the other datasets, which include: the ‘SSC,’ ‘SSC_WG,’ ‘SVIP,’ ‘SPARK’ and the ‘Denovodb’ dataset. The latter dataset was compiled by SFARI Investigator Evan Eichler and Tychele Turner and contains de novo variants from a number of neurodevelopmental, psychiatric and neurodegenerative conditions4. In addition to being searchable in the GPF platform, this dataset also has a dedicated web browser (created by Eichler and Turner).
Table 1: Datasets currently available in the Genotypes and Phenotypes in Families (GPF) platform. For each dataset, disorders for which genomics data are available, the cohort/source from which the data were derived and the type of data available (CNV: copy number variants; TG: targeted resequencing of selected genes; WES: whole-exome sequencing; WGS: whole-genome sequencing data) are summarized. The ‘Sequencing de novo’ and ‘Denovob’ datasets only include data from de novo variants. The SSC, SVIP and SPARK datasets include data from both de novo and inherited variants.
| Dataset | Disorders | Cohort/ source |
# of families |
Genomics data |
de novo mutations |
Inherited mutations |
|---|---|---|---|---|---|---|
| Sequencing de novo | autism, developmental & psychiatric disorders | SSC and published studies | 7,748 | TG, WES | Yes | No |
| Denovodb | autism, developmental, psychiatric & degenerative disorders | SSC and published studies | 11,834 | WES | Yes | No |
| SSC | autism | SSC | 2,865 | CNV, TG, WES | Yes | Yes |
| SSC_WG | autism | SSC | 510 | WGS | Yes | Yes |
| SVIP | autism & developmental disorders | Simons VIP | 84 | WES | Yes | Yes |
| SPARK | autism | SPARK | 441 | WES | Yes | Yes |
In a live demonstration, Iossifov described the platform’s functionalities. Data can be queried by dataset (e.g., ‘Sequencing de novo dataset’) using a ‘Genotype browser’ and ‘Enrichment tool.’ The former allows searching for gene variants, with options for customizing each query based on the disorder of interest, variant type, effect type (e.g., likely gene-disrupting, non-synonymous mutations), gender of the proband, as well as gene weight (e.g., the probability of loss-of-function intolerance or pLI score). Variants can be called for any number of genes as well as pre-defined gene sets, including the SFARI Gene list. Through the ‘Enrichment tool,’ investigators can also directly test whether any preexisting list of genes (e.g., from previous studies) is enriched for autism variants, as well as for variants from the other disorders available in the platform.
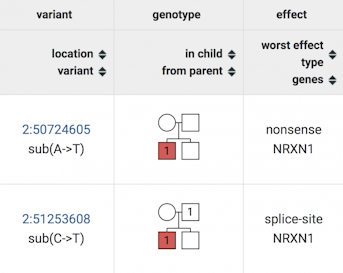
For any gene of interest, the Genotype browser returns information about variants and their distribution in the family pedigree. This example illustrates a de novo (top row) and an inherited (bottom row) variant of the NRXN1 gene, from the Simons Simplex Collection (SSC). Each variant is linked to the University of California Santa Cruz (UCSC) Genome Browser for more information. The pedigree color indicates unaffected (white) parents/siblings and children with autism (red). Circles indicate females; squares are males. For each variant, the mutation type is also shown (nonsense, splice-site).
In addition to these tools, the SSC datasets — which have phenotypic information available — can be perused using the ‘Phenotype browser’ and ‘Phenotype tool.’ Both options allow users to explore the behavioral data and the clinical and medical history of the participants; but while the ‘Phenotype browser’ allows users to search for information by the ‘instrument’ (i.e., the battery of tests used to characterize individuals in the collection), the ‘Phenotype tool’ provides details on the genetic mutations associated with any phenotypic trait and the number of individuals who carry those mutations in the collection.
Results from the Genotype browser’s searches are returned in the form of easy-to-navigate tables. For instance, queries about gene variants provide information about variant location, vulnerability/intolerance and allele frequency, as well as the pedigree of the proband (Figure 1). Illustrating how pathogenic variants segregate within the family is especially important for the study of inherited variants, which, in contrast to de novo variants, are passed down from the parent to the affected child.
Phenotypic information is summarized as distribution plots, which makes it easy to eyeball differences in the severity of a trait or the most affected gender (Figure 2). Additionally, the ‘Phenotype tool’ charts phenotypic differences between carriers and non-carriers of mutations in any gene of interest and provides a p-value of their statistical significance, which may be useful for exploring potential genotype-phenotype relationships (Figure 3).
“We created this platform to help researchers test hypotheses and interpret the SFARI datasets,” said Iossifov. “We hope this tool will facilitate perusing these large datasets, even for scientists who don’t have a background in bioinformatics.”
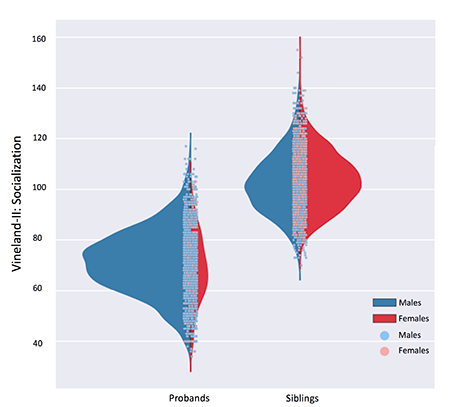
The Phenotype browser visualizes the distribution of behavioral data from Simons Simplex Collection (SSC) participants. Here, socialization scores collected through the Vineland Adaptive Behavior Scales, Second Edition (Vineland-II) are shown. Male (blue) and female (red) data are plotted separately for probands and their unaffected siblings.
Hub.iobio platform
The other genomics platform that was discussed, Hub.iobio, is based on the iobio open-source project and is currently being developed by Frameshift. Alistair Ward, president and co-founder of Frameshift, presented the functionalities and applications of Hub.iobio for autism research. The first version of the platform is expected to be released later this year and will include whole-exome and whole-genome data from the SSC, as well as whole-exome data from SPARK.
Hub.iobio differs from GPF in several respects. First, it enables users to operate on the raw sequence data directly, while GPF only allows analyzing variants that have already been identified. Second, Hub.iobio offers the possibility to visualize SFARI genotypic and phenotypic data dynamically and in real-time. For instance, samples that deviate from a normative distribution can be visually scanned and probed further. Also, subsets of data (e.g., individuals with IQ scores lower than 60) can be created with a click and then compared with the whole dataset or interrogated in relation to genetic variables of interest. Crucially, these analyses can be done interactively; researchers can explore how data distributions change by manipulating variables in real-time and quickly identify outliers and correlations — something that is not possible with a static database.
This immediate feedback will not only help accelerate the analysis of SFARI datasets, but it may also foster novel explorations of the data and the generation of original hypotheses. Whether such hypotheses will meet statistical thresholds of significance can then be assessed through standard tests.
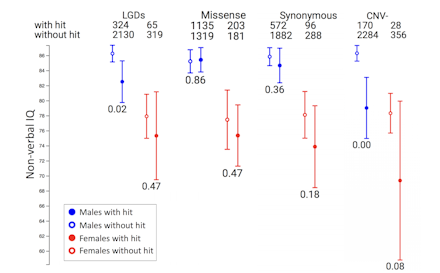
For any phenotypic measure in the collection, the Phenotype tool shows the number of individuals with and without hits [e.g., likely gene disrupting (LGD), missense and synonymous mutations, and copy number variant deletions (CNV-)]. The p-values indicate significant differences between individuals with and without hits, for each sex. In this example, Simons Simplex Collection (SSC) males with LGD mutations and CNV deletions have significantly lower non-verbal IQ than males without those mutations.
Concluding remarks
In summary, both the GPF and Hub.iobio platforms aim to facilitate data access, integration and analysis of multiple large autism cohorts. By helping to visualize genotype-phenotype relationships and extract patterns from complex genomic datasets, these tools may assist researchers in formulating novel hypotheses on how genetic mutations translate into altered behaviors.
In the near future, the developers plan to make GPF open source, so that users can install the platform on their machines and peruse their own data. New tools will be created for Hub.iobio to enable users to upload and analyze their own data. More features will also be added to analyze variants in more details (e.g., mutation type), similar to the options currently available in GPF.
“This workshop brought to light the analytical tools that are being developed to help researchers mine SFARI datasets,” said Natalia Volfovsky, director of Data and Analytics at the Simons Foundation and organizer of the workshop. “We hope these user-friendly platforms will facilitate data interpretation and advance our understanding of autism biology.”
SFARI data are accessible to approved researchers (either via the platforms described above, or users can also request the original data files via SFARI Base). More information on how to request data from the SFARI collections can be found here.