Findings from the first SPARK (Simons Foundation Powering Autism Research for Knowledge) genetics study have now been published1. The pilot study, led by the SPARK Consortium, analyzed exome and genome-wide genotyping data from 457 families with at least one offspring affected by autism spectrum disorder (ASD). Key findings include the confirmation of known ASD risk genes as well as the identification of new genes that likely confer risk for the condition.
ASD is a neurodevelopmental disorder characterized by challenges in social communication and restricted and repetitive behaviors. Genetic risk is known to contribute to the majority of ASD cases, and while over 100 genes and copy number variants (CNVs) have been linked to the disorder, many more genetic variants associated with ASD are expected to be found.
SPARK, which launched nationally in 2016, aims to recruit, engage and retain a community of 50,000 individuals with autism and their family members2. SPARK is the largest autism research cohort in the world and should provide sufficient data to power the discovery of many novel ASD risk factors.
The SPARK pilot study, which is published in npj Genomic Medicine, identified 647 rare de novo single nucleotide variants (SNVs) and indels that appeared at higher frequency in the probands compared to those without autism, including many de novo likely gene disrupting and deleterious missense variants. Many of these variants were previously known to be associated with autism, while others emerged for the first time.
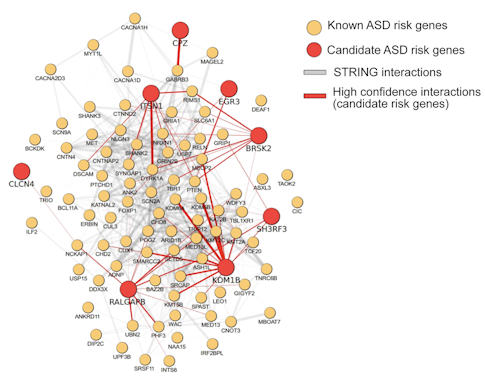
One key outcome was the discovery of a new high-confidence autism risk gene, BRSK2. Additional autism candidate genes were identified at a less stringent statistical threshold (CPZ, DMWD, FEZF2, ITSN1 and PAX5), and more evidence is needed to confirm these preliminary findings. Importantly, many of the candidate genes identified in the study function in biological pathways that have been previously implicated in autism (Figure 1) and are highly expressed during midfetal stages, a time point in brain development that has been linked to the pathogenesis of ASD3.
“We are extremely pleased with these results and are hard at work analyzing the exome and genotyping data obtained from a much larger batch of samples of approximately 6,500 individuals with autism and both their parents,” says Pamela Feliciano, SPARK scientific director and SFARI senior scientist.
To date, 36,873 individuals with autism (including 18,389 trios consisting of the individual with autism and both biological parents) have contributed DNA to SPARK. Of these, 25,000 individuals with autism will be in SPARK’s sequencing pipeline by the end of 2019, with many more thousands to be completed in the next few years.
“We expect that SPARK data will accelerate the discovery of additional genetic risk factors in autism. I am hopeful that a better understanding of the biological roots of autism will also accelerate improved outcomes for individuals with ASD,” adds Feliciano.
The SPARK team curates a list of genes and CNVs that are known to be associated with ASD; this list is routinely updated as more data accumulate in the literature. One purpose of the SPARK gene list is to identify those genes for which SPARK returns results to families — a commitment that SPARK makes to all participants who have elected to receive these results. From the pilot study, the SPARK team was able to return autism-related genetic findings to about 10 percent of the participants. An increasing number of SPARK families will receive a genetic finding in the coming months and years, as sample sizes increase and more discoveries are made.
SPARK’s growing cohort offers research opportunities to the whole autism scientific community. Genetic and phenotypic data collected by SPARK are available to approved researchers via SFARI Base. To date, this includes exome-sequencing data and genome-wide genotyping data for more than 27,000 participants (including 5,279 individuals with ASD and their parents) and phenotypic data from more than 150,000 individuals (including 59,000 with ASD). Moreover, because SPARK participants are re-contactable, researchers who would like to recruit SPARK participants for their own studies can do so via SPARK’s research match program.
“The human and scientific value of the SPARK cohort is enormous,” says Wendy Chung, director of clinical research at SFARI, Principal Investigator of SPARK, and the Kennedy Family Professor of Pediatrics in Medicine at Columbia University. “We hope SPARK will promote a better understanding of autism and lead to better supports that improve the quality of life for individuals and families with autism.”


