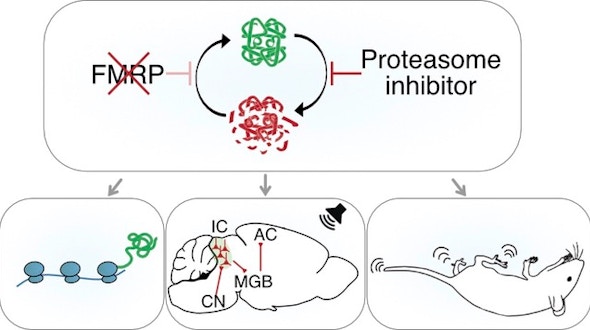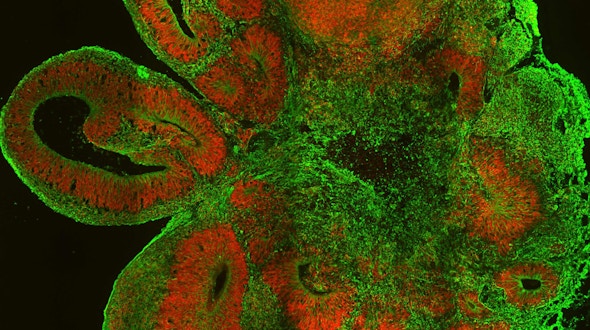
Two studies by different research groups — one led by Flora Vaccarino and the other by Juergen Knoblich — used brain organoids derived from pluripotent stem cells of people with autism and showed how transcriptional alterations affecting certain cell types during human brain development could contribute to the early emergence of ASD.